FTDMP
FTDMP (Flexible Toolkit Dedicated to Multimeric Predictions)
FTDMP is a framework for protein-protein, protein-DNA and protein-RNA docking and scoring. It can be used for biomolecular docking and subsequent scoring, or just for scoring and ranking structural models of biomolecular complexes generated by different structure prediction methods.
The software is available on GitHub, together with it's documentation, including installation and usage instructions.
FTDMP has been tested using protein-protein, protein-DNA and protein-RNA docking benchmarks. The benchmark datasets are available at Zenodo together with docking tables. FTDMP docking results for the benchmarks are also available at Zenodo.
Publications
If you use the FTDMP for your research, please cite the following articles.
FTDMP software, cleaned docking benchmarks and docking results are published here:
- Olechnovič K, Banciul R, Dapkūnas J, Venclovas Č. (2025) FTDMP: A Framework for Protein-Protein, Protein-DNA, and Protein-RNA Docking and Scoring. Proteins. doi: 10.1002/prot.26792. PubMed PMID: 39748638.
Scoring of protein-protein interfaces using the VoroIF-jury algorithm and details of this algorithm are published in our CASP15 article:
- Olechnovič K, Valančauskas L, Dapkūnas J, Venclovas Č. (2023) Prediction of protein assemblies by structure sampling followed by interface-focused scoring. Proteins; 91:1724–1733. doi: 10.1002/prot.26569. PubMed PMID: 37578163.
VoroMQA
VoroMQA: assessment of protein structure quality using interatomic contact areas
VoroMQA web-server is available at bioinformatics.ibt.lt/wtsam/voromqa.
VoroMQA software for Linux is included in the latest Voronota package available from bitbucket.org/kliment/voronota/downloads.
Testing data tables are available here.
Sector of Microtechnologies
Collaborations Prof. Allon M. Klein (Harvard Medical School, USA) https://sysbio.med.harvard.edu/facultys/allon-klein-phd Ongoing Projects: H2020-MSCA-ITN-2018, EvoDrops (813786), directed EVOlution in DROPS 01.2.2-MITA-K-702 "Single genome amplification of clinically relevant microorganisms using microfluidics technology", EUREKA Network E! 13634 SAGCHIP 01.2.2-LMT-K-718-04-0002, ”Establishment of single-cell transcriptomics/genomics research parallel-laboratory" 09.3.3O-LMT-K-712-01-0056, "Droplet microfluidics for single-cell genotype phenotype linkage research" |
Linas Mazutis, Ph.D., Prof. Lab Members: Graduate Students: |
Visit our new group's webpage: https://www.gmc.vu.lt/en/sector-of-microtechnologies
Research Overview
Droplet microfluidics technology provides a powerful tool in life sciences. The basic principle of this technology is easy to appreciate: highly monodisperse aqueous droplets are generated in an inert carrier oil in microfluidic channels on a chip and each droplet functions as an independent microreactor. Hence, each droplet is the functional equivalent of a well (or tube), yet the volume of droplet is roughly a thousand to a million times smaller. Such massive reduction in reaction volume provides huge savings in reagents cost when performing large numbers of reactions in parallel. Furthermore, unlike the conventional microtiter plates or valve-based microfluidics, droplets are intrinsically scalable: the number of reaction ‘wells’ is not limited by the physical dimensions of the chip but scales linearly with the emulsion volume. Different microfluidic modules can be employed to manipulate droplets in sophisticated, yet highly controllable manner. Large numbers of droplets (>10^9) can be generated at astonishingly high rates (>20,000 droplets per second), their size tuned precisely, new reagents introduced into pre-formed droplets at defined time points, droplet split and sorted, therefore opening new opportunities for single-cell -omics field. Many useful microfluidic techniques have been developed to profile and even selectively purify single-cells, however, the demand for methods with better analytical performance and improved high-throughput capabilities, remains very high. We are working at fulfilling this demand by bringing higher throughput, reduced reagent cost, scalability and single-molecule resolution for diverse set of quantitative experiments in cell biology and biomedicine.
1. Single-cell barcoding and sequencing using droplet microfluidics
Single-cell sequencing technologies has affected all branches of biological and biomedical sciences, yet one of the major obstacles imposing a real breakthrough is relatively low number of cells that can be isolated and sequenced. Deciphering the unbiased composition of heterogeneous cell populations requires innovative techniques that could capture not hundreds but thousands of single-cells and to do so in a high-throughput manner and affordable cost. This work is built on idea of isolating individual cells into microfluidic droplets carrying RNA/DNA barcodes, and assay reagents. Because individual cells are compartmentalized in drops, cell genetic make up and contents can be accessed. For single-cell studies our approach provides unprecedented scalability, significantly increased throughput, minimal sample loss, reagent savings and large experimental flexibility. Using this technology we are interested in understanding the genetic and epigenetic factors that are responsible for cellular heterogeneity and inheritance.
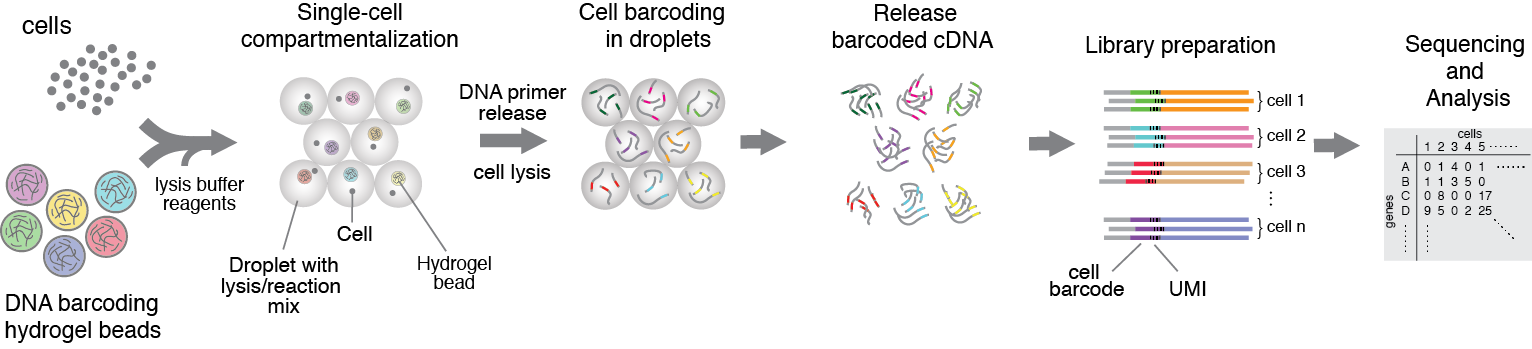
Figure 1. Heterogenious mix of cells is encapsulated into droplets together with reverse-transcription enzyme, lysis mix, and hydrogel beads carrying barcoding primers. After encapsulation cells are lysed, primers are released from the beads and cDNA is tagged with a barcode during reverse transcription reaction. Once cDNA synthesis is complete, droplets are broken and barcoded material from all cells is amplified for next-gen sequencing. Read to learn more
Zilionis R., Nainys J, Veres A., Savova V., Zemmour D., Klein MA., and Mazutis L, (2017) Single-cell barcoding and sequencing using droplet microfluidics, Nature Protocols 12, 44–73
Klein M*, Mazutis L*, Akartuna I*, Tallapragada N, Veres A, Li V, Peshkin L, Weitz D and Kirschner M, (2015) Droplet barcoding for single cell transcriptomics applied to embryonic stem cells, Cell, 161(5): 1187–1201
* - joint first co-authors, http://www.cell.com/cell/first-reflections/mazutis
2. High-throughput screening of antibody secreting cells
In this project, we use droplet microfluidics platform for high-throughput single-cell screening of cells that produce therapeutic antibodies or biomolecules of biomedical interest. Cell compartmentalization into microfluidic droplets together with capture beads and barcoded DNA primers enables us a direct establishment of the linkage between the genotype (genes or mRNA) and phenotype (binding, regulatory or activity of secreted proteins). Due to increased local concentration inside droplets the amount of secreted antibodies will significantly reduce the time window necessary for the assay. We aim at developing a discovery pipeline for the quantitative high-throughput antibody phenotyping without loosing the original heavy-light chain pairing, which would be a significant advantage over other technologies. Our technological approach provides a unique way to identify the primary sequence of heavy and light IgG genes encoding functional monoclonal antibodies directly from single-cells, without a need to perform gene cloning, hybridoma construction or cell immortalization.
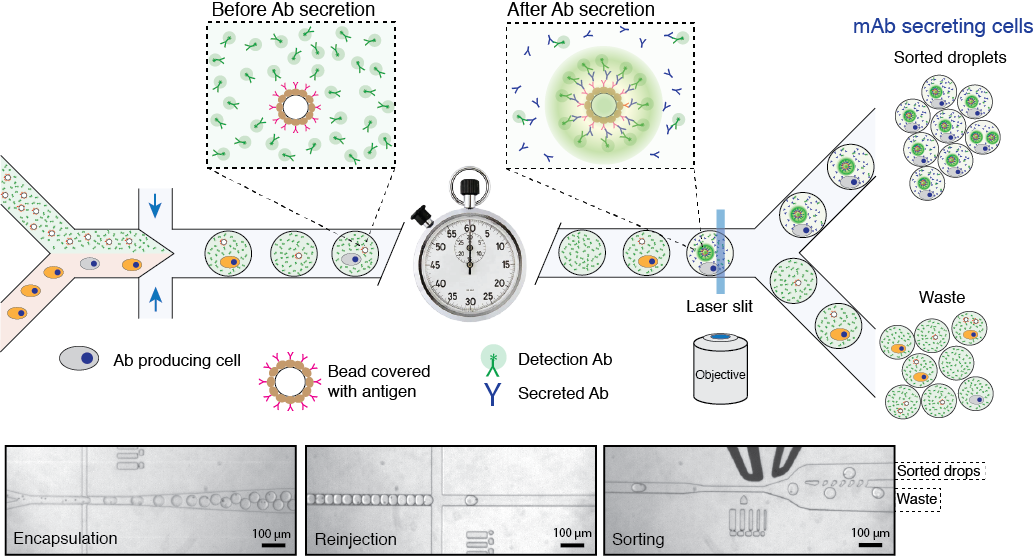
Figure 2. Schematics of microfluidics platform for screening and sequencing single-cells. A mixture of cells is loaded into droplets together with assay reagents and beads carrying capture probe (antigen). The cells secreting antigen specific Ab will be detected by fluorescent sandwich assay on bead surface and sorted. The sorted cells will be lysed and mRNA encoding heavy and light chains are captured and sequenced. Alternatively, the sorted cells can be recovered directly into a growth media for further cultivation, propagation and analysis. Read to learn more
Mazutis L., Gilbert J., Ung L., Weitz D., Griffiths A., Heyman J., (2013) Single-cell analysis and sorting using droplet-based microfluidics, Nature Protocols, 8(5): 870-891.
3. in vitro directed evolution using artificial cells
Darwinian evolution is a powerful algorithm that has given rise to the functionally diverse set of proteins present in all living systems. Repetitive rounds of mutation, selection and amplification have optimized nature’s catalysts, the enzymes, to an extraordinary degree, for example. Biocatalysts are typically much more efficient than their man-made counterparts, often working close to the diffusion limit. A better understanding of how new enzymes evolve consequently remains an important and challenging task for both academic and industrial needs. Although the active sites of natural enzymes are highly complex, making the design of new or promiscuous protein functions difficult, computation has recently emerged as a potentially powerful strategy for creating protein catalysts with tailored activities and specificities. Because the activities of such artificial enzymes are still modest, we plan to explore the feasibility of optimizing them by combining the latest advances in droplet-based microfluidics technology with the methods of directed evolution. Specifically, two complimentary groups, one with expertise in microfluidic systems and another with expertise in directed protein evolution, will join forcess to investigate mechanisms and strategies for optimizing several computationally designed esterases. By applying droplet-based microfluidics technology, in which biochemical reactions are performed inside micrometer size vessels, we will have a powerful tool enabling large number of libraries (>10^6) to be screened at ultra-high-throughput rates and using conditions that are incompatible with in vivo systems.
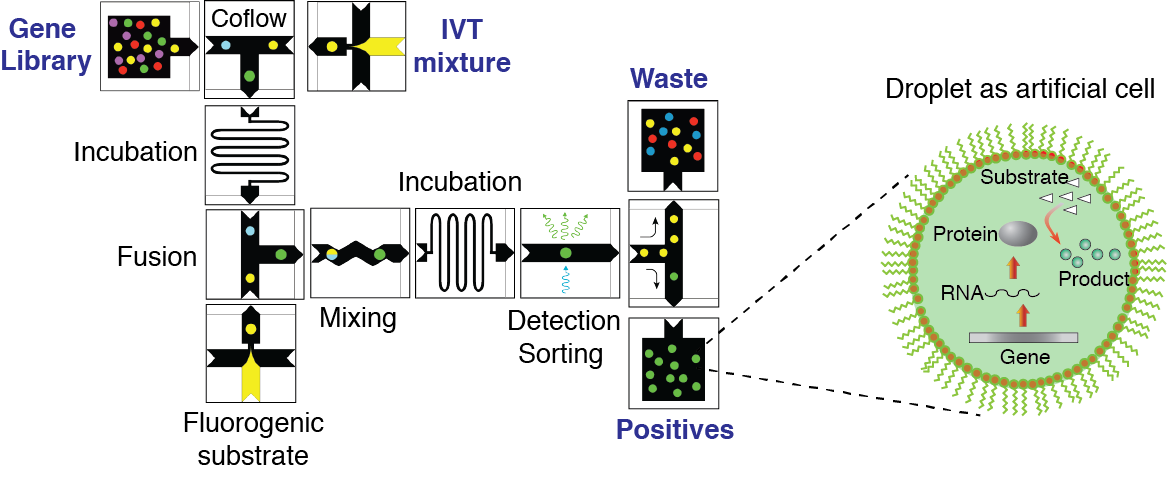
Figure 3. Concept of directed evolution approach in droplet microfluidics. Droplets containing single-genes with all ingredients necessary for in vitro expression will serve as artificial cells that can be selected for a desirable phenotype under conditions that are not feasible in living systems. Library of mutated genes is encapsulated such that, statistically, the majority of droplets contain no more than one gene per droplet. After IVTT reaction the droplets are fused with a second type of droplet containing a fluorogenic substrate and other molecules, salts, and reagents needed to impose the selection pressure. After mixing and incubation, the fluorescence of each droplet is recorded. Droplets with fluorescence above a threshold are sorted by means of dielectrophoresis and the genes contained therein are amplified using PCR. The selected genes can be either characterized, re-selected, or mutated for subsequent round of directed evolution.
Galinis R, Stonyte G, Kiseliovas V, Zilionis R, Studer S, Hilvert D, Janulaitis A and Mazutis L. (2016) DNA nanoparticles for improved protein synthesis in vitro, Angew Chem Int Ed Engl. 2016 Feb 24;55(9):3120-3
4. Microfluidic tools for biological and biomedical applications
Our group has long history of experience at developing novel microfluidic tools for precise manipulation and analysis of biological reactions at pico and nanoliter range volumes. We provide designs, modeling, fabrication and characterization of microfluidic systems and devices for myriad of biological and biomedical applications. We build experimental platforms based on integration of microfluidics, mechanical, electrical and optical modules eventually enabling precise control over experimental conditions hardly feasible in bulk. Using droplet microfluidic devices we provide significantly reduced assay volumes for virtually any biological assay and as a result big cost savings for biochemical reagents and compounds. We are developing microfluidic devices and chips for digital DNA/RNA analysis, single-cell and genomic applications. In addition to droplet microfluidic systems in collaboration with Dr. Jonathan Thon and Prof. Joseph Italiano (Brigham and Women's Hospital, USA) we have developed a micro-bioreactor for generation of synthetic human platelets in vitro. This work formed a technological base for establishment of a start-up company Platelet Biogenesis Inc., which is based in Boston, MA.
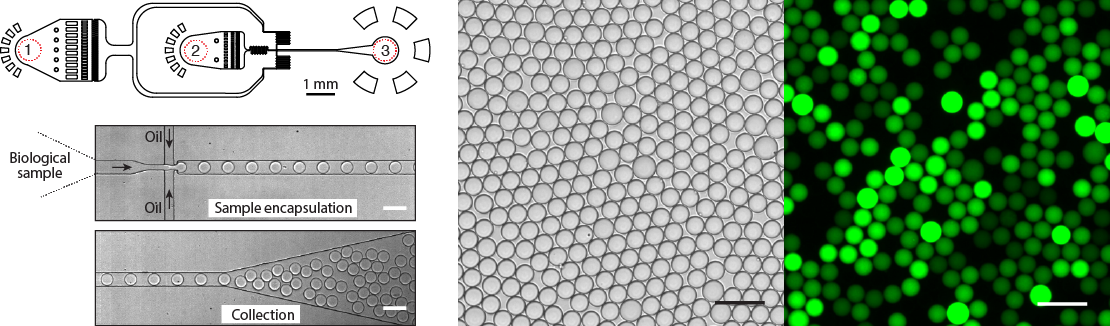
Figure 4. Design and operation of the droplet microfluidics device. Almost any biological sample (DNA, cells, proteins) can be encapsulated into pico- or nano-liter volume droplets at >10.000 droplets per second. Typical microfluidics device has (1) the inlet for the continuous phase, (2) the inlet for biological sample, and (3) the droplet collection outlet. Bright field and fluorescence images of an emulsion after DNA amplification reaction. Droplets containing amplified DNA exhibit green fluorescence, whereas droplets lacking a template are dark. Scale bars, 50 um.
Mazutis L, Araghi AF, Miller OJ, Baret JC, Frenz L, Janoshazi A, Taly V, Miller BJ, Hutchison JB, Link D, Griffiths AD, Ryckelynck M., (2009) Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis, Analytical Chemistry 81(12): 4813-21.
Thon JN, Mazutis L, Wu S, Sylman JL, Ehrlicher A, Machlus KR, Feng Q, Lu S, Lanza R, Neeves KB, Weitz DA, Italiano JE (2014) Platelet bioreactor-on-a-chip, Blood, 124(12): 1857-1867
Mazutis L. and A. Griffiths (2012) “Selective droplet coalescence using microfluidic systems”. Lab on a Chip, 12, 1800-1806.
5. Drug delivery systems
Vesicles and microspheres composed of biodegradable polymers are becoming increasingly attractive agents for improved delivery, stabilization and prolonged release of drugs. However, the vast majority of techniques being used for microspheres production are based on mechanical steering or sonication. These methods, although efficient, produce particles with large polydispersity (coefficient of variance C.V. ~10-30%) and therefore impose several limitations. Particle’s size distribution may produce uncontrolled variation in the rate of particle degradation, affect the kinetics of drug release and decrease the stability of encapsulated drugs. In contrast, monodisperse drug-loaded particles offer significantly better control over release kinetics and exhibit a much lower initial drug release burst that is observed when using polydisperse particles. We aim to develop droplet-based microfluidics technology that will be used to create highly monodisperse (C.V. < 0.5%) vesicles and particles for improved drug encapsulation, delivery and release. Microfluidic systems offer a novel way to create and manipulate fluids that is hardly possible using conventional techniques. For instance, miniaturized microfluidic systems have been used to create polymer particles composed of thermosensitive, mechanosensitive, ion-sensitive and biodegradable materials, finding many useful applications in biotechnology and chemical industry. In this project we will develop and apply microfluidic technology for production of nano- and pico-liter volume particles in which inner part of the vesicle will contain preloaded biological or chemical molecules and outer part (shell) will be composed of biodegradable polymer. Additional advantage of using microfluidics is that inner and outer parts of the vesicles can be tuned by controlling the volumetric flow rate ratio, thus allowing precise control over the vesicle size and thickness of the shell.
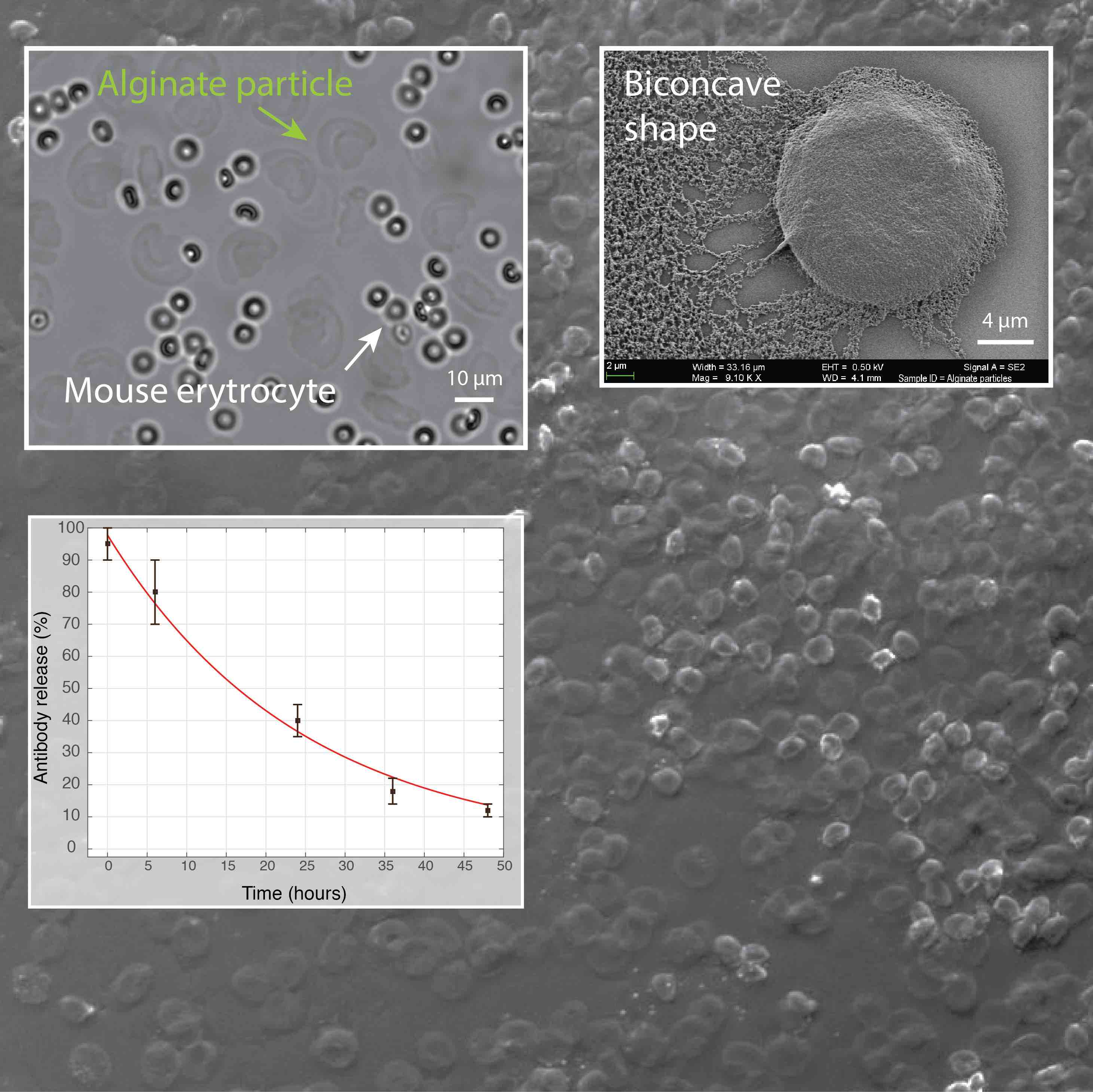
Figure 5. We have developed a microfluidic approach for production of hydrogel particles having the unique biconcave shape and the size of a mammalian cell (~10 µm). The release of encapsulated antibodies was largely affected by the presence of phosphate ions that chelate calcium (a cross-linker of alginate polymer) and cause the dissolution of hydrogel. In addition, the image shows the biocompatibility of alginate particles in the whole blood.
Vasiliauskas R, Liu D, Cito S, Zhang H, Shahbazi MA, Sikanen T, Mazutis L, Santos HA (2015) A Simple Microfluidic Approach to Fabricate Monodisperse Hollow Microparticles for Multidrug Delivery, ACS Appl. Mater. Interfaces, 7 (27), pp 14822–14832.
Mazutis L, Vasiliauskas R, Weitz D, (2015) Microfluidic Production of Alginate Hydrogel Particles for Antibody Encapsulation and Release, Macromol. Biosci. 2015, 15, 1641–1646
6. DNA-Magnesium-pyrophosphate particle biosynthesis and their use
Compartmentalization and amplification of single DNA molecules inside nano- or pico-liter sized wells and droplets opens new opportunities for biomedical and biological sciences. The most common method of amplifying DNA in a sample involves the polymerase chain reaction (PCR). Yet, amplification of long (>1 kb) single-molecule templates is often inefficient, leading to reduced reaction yields and biases. DNA amplification under isothermal reaction conditions has been shown to generate large amounts of material from a single-copy DNA template. Moreover, the ability to amplify DNA and then express proteins from the clonally amplified template would greatly increase the scope of potential applications.
We employ droplet microfluidics devices to encapsulate and convert single DNA molecules into condensed DNA nanoparticles by a multiple displacement amplification (MDA) reaction driven by phi29 DNA polymerase. The condensed DNA nanoparticles comprise up to ~10.000-100.000 copies of clonally amplified DNA template. Intriguingly, we found that inorganic pyrophosphate, produced during isothermal DNA synthesis, and magnesium ions are a prerequisite for DNA condensation into the crystalline-like globular structures. This process is enhanced when the DNA amplification reaction is performed inside droplets, which can be attributed to the confined volumes and spatial accumulation of the reaction products. We have demonstrated the biological functionality of the DNA nanoparticles, by applying them in in vitro transcription-translation reactions and observed improved protein expression yields relative to standard assay conditions.
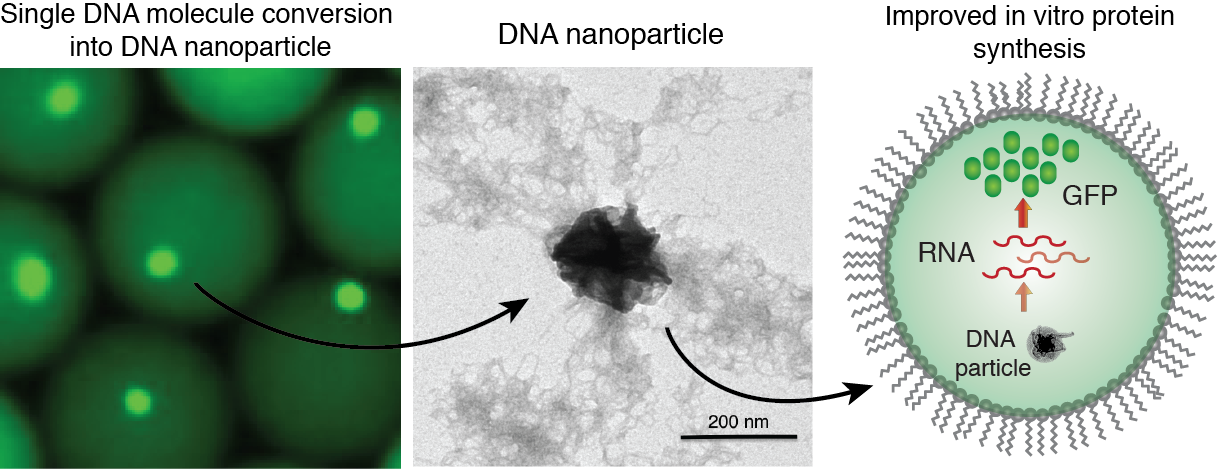
Figure 6. Generation of DNA-Mg-PP particles. DNA nanoparticle formation induced by inorganic pyrophosphate (PP) and magnesium ions during a phi29-catalyzed DNA polymerization reaction. TEM image of the DNA-Mg-PP particle. The use of DNA for efficient gene expression and protein synthesis in vitro. Read to learn more
Galinis R, Stonyte G, Kiseliovas V, Zilionis R, Studer S, Hilvert D, Janulaitis A and Mazutis L. (2016) DNA nanoparticles for improved protein synthesis in vitro, Angew Chem Int Ed Engl. 2016 Feb 24;55(9):3120-3
Zubaite, G.; Simutis, K.; Galinis, R.; Milkus, V.; Kiseliovas, V.; Mazutis, L. Droplet Microfluidics Approach for Single-DNA Molecule Amplification and Condensation into DNA-Magnesium-Pyrophosphate Particles. Micromachines 2017, 8, 62.
Funding sources
European Commission, H2020 program http://ec.europa.eu/research/mariecurieactions/
Lithuanian-Swiss Research and Development cooperation program https://www.cpva.lt/en/lithuanian-swiss-cooperation-programme
The Scientific Exchange Programme (Sciex-NMSch) https://www.swissuniversities.ch/en/topics/sciex/
Agency for Science, Innovation and Technology (MITA) http://www.mita.lt/en
Lithuanian Research Council http://www.lmt.lt/en/about.html
Important publications of our group:
1. Klein M*, Mazutis L*, Akartuna I*, Tallapragada N, Veres A, Li V, Peshkin L, Weitz D and Kirschner M, (2015) Droplet barcoding for single cell transcriptomics applied to embryonic stem cells, Cell, 161(5): 1187–1201
2. Zilionis R., Nainys J, Veres A., Savova V., Zemmour D., Klein MA., and Mazutis L, (2017) Single-cell barcoding and sequencing using droplet microfluidics, Nature Protocols 12, 44–73
3. Azizi E, Carr A, Plitas G., Cornish EA, Konopacki C, Prabhakaran S, Nainys J., Wu K. Kiseliovas V., Setty M., Choi K., Dao P, Mazutis L, Rudensky YA & Peer D, Single-cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment (2018), Cell 174, 1–16, August 23
4. Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe'er D., (2019) Characterization of cell fate probabilities in single-cell data with Palantir, Nat Biotechnol. 2019 Apr;37(4):451-460.
5. Chi, Y., J. Remsik, V. Kiseliovas, C. Derderian, U. Sener, M. Alghader, F. Saadeh, K. Nikishina, T. Bale, C. Iacobuzio-Donahue, T. Thomas, D. Pe'er, L. Mazutis and A. Boire (2020). "Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis." Science 369(6501): 276-282.
Hot publications:
1. Pritykin, Y., J. van der Veeken, A. R. Pine, Y. Zhong, M. Sahin, L. Mazutis, D. Pe'er, A. Y. Rudensky and C. S. Leslie (2021). "A unified atlas of CD8 T cell dysfunctional states in cancer and infection." Mol Cell 3;81(11):2477-2493
2. Alonso-Curbelo, D., Y. J. Ho, C. Burdziak, J. L. V. Maag, J. P. t. Morris, R. Chandwani, H. A. Chen, K. M. Tsanov, F. M. Barriga, W. Luan, N. Tasdemir, G. Livshits, E. Azizi, J. Chun, J. E. Wilkinson, L. Mazutis, S. D. Leach, R. Koche, D. Pe'er and S. W. Lowe (2021). "A gene-environment-induced epigenetic program initiates tumorigenesis." Nature 590(7847): 642-648.
3. Leonaviciene, G., Leonavicius, K., Meskys, R. & Mazutis, L. Multi-step processing of single cells using semi-permeable capsules. Lab on a Chip 20, 4052-4062 (2020).
Publications:
2017-present
Pritykin, Y., J. van der Veeken, A. R. Pine, Y. Zhong, M. Sahin, L. Mazutis, D. Pe'er, A. Y. Rudensky and C. S. Leslie (2021). "A unified atlas of CD8 T cell dysfunctional states in cancer and infection." Mol Cell 3;81(11):2477-2493
Guttikonda, S. R., L. Sikkema, J. Tchieu, N. Saurat, R. M. Walsh, O. Harschnitz, G. Ciceri, M. Sneeboer, L. Mazutis, M. Setty, P. Zumbo, D. Betel, L. D. de Witte, D. Pe'er and L. Studer (2021). "Fully defined human pluripotent stem cell-derived microglia and tri-culture system model C3 production in Alzheimer's disease." Nat Neurosci 24(3): 343-354.
Alonso-Curbelo, D., Y. J. Ho, C. Burdziak, J. L. V. Maag, J. P. t. Morris, R. Chandwani, H. A. Chen, K. M. Tsanov, F. M. Barriga, W. Luan, N. Tasdemir, G. Livshits, E. Azizi, J. Chun, J. E. Wilkinson, L. Mazutis, S. D. Leach, R. Koche, D. Pe'er and S. W. Lowe (2021). "A gene-environment-induced epigenetic program initiates tumorigenesis." Nature 590(7847): 642-648.
2020
The Human Tumor Atlas Network: Charting Tumor Transitions across Space and Time at Single-Cell Resolution. Rozenblatt-Rosen O, Regev A, Oberdoerffer P, Nawy T, Hupalowska A, Rood JE, Ashenberg O, Cerami E, Coffey RJ, Demir E, Ding L, Esplin ED, Ford JM, Goecks J, Ghosh S, Gray JW, Guinney J, Hanlon SE, Hughes SK, Hwang ES, Iacobuzio-Donahue CA, Jané-Valbuena J, Johnson BE, Lau KS, Lively T, Mazzilli SA, Pe'er D, Santagata S, Shalek AK, Schapiro D, Snyder MP, Sorger PK, Spira AE, Srivastava S, Tan K, West RB, Williams EH; Human Tumor Atlas Network. Cell. 2020 Apr 16;181(2):236-249.
Laughney AM, Hu J, Campbell NR, Bakhoum SF, Setty M, Lavallée VP, Xie Y, Masilionis I, Carr AJ, Kottapalli S, Allaj V, Mattar M, Rekhtman N, Xavier JB, Mazutis L, Poirier JT, Rudin CM, Pe'er D, Massagué J. Regenerative lineages and immune-mediated pruning in lung cancer metastasis. Nat Med. 2020 Feb;26(2):259-269.
Leonaviciene, G., K. Leonavicius, R. Meskys and L. Mazutis (2020). "Multi-step processing of single cells using semi-permeable capsules." Lab Chip 20(21): 4052-4062.
Gegevicius, E., Goda, K., and Mazutis, L. (2020). CHAPTER 4:Droplet Gene Analysis – Digital PCR. In Droplet Microfluidics, pp. 89–121. DOI: 10.1039/9781839162855-00089
Chi, Y., J. Remsik, V. Kiseliovas, C. Derderian, U. Sener, M. Alghader, F. Saadeh, K. Nikishina, T. Bale, C. Iacobuzio-Donahue, T. Thomas, D. Pe'er, L. Mazutis and A. Boire (2020). "Cancer cells deploy lipocalin-2 to collect limiting iron in leptomeningeal metastasis." Science 369(6501): 276-282.
Ding, R. H., K. C. Hung, A. Mitra, L. W. Ung, D. Lightwood, R. Tu, D. Starkie, L. H. Cai, L. Mazutis, S. R. Chong, D. A. Weitz and J. A. Heyman (2020). "Rapid isolation of antigen-specific B-cells using droplet microfluidics." RSC Advances 10(45): 27006-27013.
Karthaus, W. R., M. Hofree, D. Choi, E. L. Linton, M. Turkekul, A. Bejnood, B. Carver, A. Gopalan, W. Abida, V. Laudone, M. Biton, O. Chaudhary, T. H. Xu, I. Masilionis, K. Manova, L. Mazutis, D. Pe'er, A. Regev and C. L. Sawyers (2020). "Regenerative potential of prostate luminal cells revealed by single-cell analysis." Science 368(6490): 497.
Kim, S. H., A. Holland, A. Jimenez-Sanchez, Y. Bykov, R. M. Fromme, A. Stylianou, T. Walther, C. Liu, M. M. Leitao, O. Zivanovic, Y. Sonoda, D. S. Chi, N. R. Abu-Rustum, L. Mazutis, G. Plitas, T. J. Hollmann, B. Weigelt, D. Pe'er and D. Zamarin (2020). "Compositional and architectural characterization of high-grade serous ovarian carcinomas using single cell technologies and multiplex microscopy." Gynecologic Oncology 159: 44-44.
Marjanovic, N. D., M. Hofree, J. E. Chan, D. Canner, K. Wu, M. Trakala, G. G. Hartmann, O. C. Smith, J. Y. Kim, K. V. Evans, A. Hudson, O. Ashenberg, C. B. M. Porter, A. Bejnood, A. Subramanian, K. Pitter, Y. Yan, T. Delorey, D. R. Phillips, N. Shah, O. Chaudhary, A. Tsankov, T. Hollmann, N. Rekhtman, P. P. Massion, J. T. Poirier, L. Mazutis, R. Li, J. H. Lee, A. Amon, C. M. Rudin, T. Jacks, A. Regev and T. Tammela (2020). "Emergence of a High-Plasticity Cell State during Lung Cancer Evolution." Cancer Cell 38(2): 229-246 e213.
Morello, F., D. Borshagovski, M. Survila, L. Tikker, S. Sadik-Ogli, A. Kirjavainen, N. Estartus, L. Knaapi, L. Lahti, P. Toronen, L. Mazutis, A. Delogu, M. Salminen, K. Achim and J. Partanen (2020). "Molecular Fingerprint and Developmental Regulation of the Tegmental GABAergic and Glutamatergic Neurons Derived from the Anterior Hindbrain." Cell Rep 33(2): 108268.
Xue, J. Y., Y. L. Zhao, J. Aronowitz, T. T. Mai, A. Vides, B. Qeriqi, D. Kim, C. C. Li, E. de Stanchina, L. Mazutis, D. Risso and P. Lito (2020). "Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition." Nature 577(7790): 421-425.
2019
Cai XC, Zhang T, Kim EJ, Jiang M, Wang K, Wang J, Chen S, Zhang N, Wu H, Li F, Dela Seña CC, Zeng H, Vivcharuk V, Niu X, Zheng W, Lee JP, Chen Y, Barsyte D, Szewczyk M, Hajian T, Ibáñez G, Dong A, Dombrovski L, Zhang Z, Deng H, Min J, Arrowsmith CH, Mazutis L, Shi L, Vedadi M, Brown PJ, Xiang J, Qin LX, Xu W, Luo M. A chemical probe of CARM1 alters epigenetic plasticity against breast cancer cell invasion. eLife. 2019 Oct 28;8:e47110.
Brown CC, Gudjonson H, Pritykin Y, Deep D, Lavallée VP, Mendoza A, Fromme R, Mazutis L, Ariyan C, Leslie C, Pe'er D, Rudensky AY. Transcriptional Basis of Mouse and Human Dendritic Cell Heterogeneity. Cell. 2019 Oct 31;179(4):846-863.e24.
Setty M, Kiseliovas V, Levine J, Gayoso A, Mazutis L, Pe'er D., (2019) Characterization of cell fate probabilities in single-cell data with Palantir, Nat Biotechnol. 2019 Apr;37(4):451-460.
van der Veeken J, Zhong Y, Sharma R, Mazutis L, Dao P, Pe'er D, Leslie CS, Rudensky AY. (2019) Natural Genetic Variation Reveals Key Features of Epigenetic and Transcriptional Memory in Virus-Specific CD8 T Cells, Immunity. 2019 May 21;50(5):1202-1217
Nainys, Juozas, Valdemaras Milkus, and Linas Mažutis. "Single-cell screening using microfluidic systems." Microfluidics for Pharmaceutical Applications. William Andrew Publishing, 2019. 353-367.
Karolis S, Stonyte G, and Mažutis L. "Antibody discovery using microfluidic systems." Microfluidics for Pharmaceutical Applications. William Andrew Publishing, 2019. 337-351.
2018
X Fan, B Moltedo, A Mendoza, AN Davydov, MB Faire, L Mazutis, R Sharma, D Pe’er, DM Chudakov, AY Rudensky, CD49b defines functionally mature Treg cells that survey skin and vascular tissues (2018), Journal of Experimental Medicine 215 (11), 2796-2814
K Leonavicius, J Nainys, D Kuciauskas, L Mazutis, Multi-omics at single-cell resolution: comparison of experimental and data fusion approaches, (2019) Current Opinion in Biotechnology 55, 159-166
Azizi E, Carr A, Plitas G., Cornish EA, Konopacki C, Prabhakaran S, Nainys J., Wu K. Kiseliovas V., Setty M., Choi K., Dao P, Mazutis L, Rudensky YA & Peer D, Single-cell Map of Diverse Immune Phenotypes in the Breast Tumor Microenvironment (2018), Cell 174, 1–16, August 23
van Dijk D, Sharma R, Nainys J, Yim K, Kathail P, Carr A., Burdziak C., Moon RK,Chaffer C., Pattabiraman D, Bierie B, Mazutis L., Wolf G., Krishnaswamy S., Peer D.
Recovering gene interactions from single-cell data using data diffusion, (2018) Cell, 174, 716-729, July 29
2010-2017
Zilionis R., Nainys J, Veres A., Savova V., Zemmour D., Klein MA., and Mazutis L, (2017) Single-cell barcoding and sequencing using droplet microfluidics, Nature Protocols 12, 44–73
Einav T., Mazutis L, and Phillips R, (2016) Statistical Mechanics of Allosteric Enzymes, J. Phys. Chem. B, 120 (26), pp 6021–6037.
Galinis R, Stonyte G, Kiseliovas V, Zilionis R, Studer S, Hilvert D, Janulaitis A and Mazutis L. (2016) DNA nanoparticles for improved protein synthesis in vitro, Angew. Chem. Int Ed, 55, 3120-3123
Wagner O, Thiele J, Weinhart M, Mazutis L, Weitz D, Huck W Haag R. (2016) Biocompatible fluorinated polyglycerols for droplet microfluidics as an alternative to PEG- based copolymer surfactants, Lab on a Chip, 16, 65-69
Mazutis L, Vasiliauskas R, Weitz D, (2015) Microfluidic Production of Alginate Hydrogel Particles for Antibody Encapsulation and Release, Macromol. Biosciences, 15, 1641–1646
Vasiliauskas R, Liu D, Cito S, Zhang H, Shahbazi MA, Sikanen T, Mazutis L, Santos HA (2015) A Simple Microfluidic Approach to Fabricate Monodisperse Hollow Microparticles for Multidrug Delivery, ACS Appl. Mater. Interfaces, 7 (27), pp 14822–14832.
Bender M, Thon JN, Ehrlicher AJ, Wu S, Mazutis L, Deschmann E, Sola-Visner M, Italiano JE Jr, Hartwig JH. (2015) Dynein-dependent microtubule sliding drives proplatelet elongation, Journal of Thrombosis and Haemostasis, 13:184.
Klein M*, Mazutis L*, Akartuna I*, Tallapragada N, Veres A, Li V, Peshkin L, Weitz D and Kirschner M, (2015) Droplet barcoding for single cell transcriptomics applied to embryonic stem cells, Cell, 161(5): 1187–1201 * - joint first co-authors,
Bender M, Thon JN, Ehrlicher AJ, Wu S, Mazutis L, Deschmann E, Sola-Visner M, Italiano JE Jr, Hartwig JH. (2015) Microtubule sliding drives proplatelet elongation and is dependent on cytoplasmic dynein, Blood, 125(5): 860-868
Shekhar S., L. Zhu, L. Mazutis, A. E. Sgro, T. G. Fai, and M. Podolski (2014) Quantitative biology: where modern biology meets physical sciences, Molecular Biology of the Cell, Nov.5, vol.25, no.22, 3482.
Thon JN, Mazutis L, Wu S, Sylman JL, Ehrlicher A, Machlus KR, Feng Q, Lu S, Lanza R, Neeves KB, Weitz DA, Italiano JE (2014) Platelet bioreactor-on-a-chip, Blood, 124(12): 1857-1867
Mazutis L., Gilbert J., Ung L., Weitz D., Griffiths A., Heyman J., (2013) Single-cell analysis and sorting using droplet-based microfluidics, Nature Protocols, 8(5): 870-891
Mazutis L. and A. Griffiths (2012) Selective droplet coalescence using microfluidic systems, Lab on a Chip, 12, 1800-1806.
Skhiri, Y.; Gruner, P.; Semin, B.; Brosseau, Q.; Pekin, D.; Mazutis, L.; Goust, V.; Kleinschmidt, F.; El Harrak, A.; Hutchison, J. B.; Mayot, E.; Bartolo, J. F.; Griffiths, A. D.; Taly, V.; Baret, J. C., (2012) Dynamics of molecular transport by surfactants in emulsions, Soft Matter, 8 (41), 10618-10627.
Pekin, D.; Skhiri, Y.; Baret, J. C.; Le Corre, D.; Mazutis, L.; Salem, C. B.; Millot, F.; El Harrak, A.; Hutchison, J. B.; Larson, J. W.; Link, D. R.; Laurent-Puig, P.; Griffiths, A. D.; Taly, V., (2011) Quantitative and sensitive detection of rare mutations using droplet-based microfluidics. Lab Chip, 11(13), 2156-66.
Until 2010
Mazutis L. and A. D. Griffiths (2009) Preparation of monodisperse emulsions by hydrodynamic size fractionation, Appl. Phys. Lett. 95(20): 204103.
Mazutis L, Baret JC, Treacy P, Skhiri Y, Araghi AF, Ryckelynck M, Taly V, Griffiths AD. (2009) Multi-step microfluidic droplet processing: kinetic analysis of an in vitro translated enzyme, Lab on a Chip 9(20): 2902-8.
Mazutis L, Baret JC, Griffiths AD (2009) A fast and efficient microfluidic system for highly selective one-to-one droplet fusion, Lab Chip 9(18): 2665-72
Mazutis L, Araghi AF, Miller OJ, Baret JC, Frenz L, Janoshazi A, Taly V, Miller BJ, Hutchison JB, Link D, Griffiths AD, Ryckelynck M. (2009) Droplet-based microfluidic systems for high-throughput single DNA molecule isothermal amplification and analysis, Analytical Chemistry 81(12): 4813-21.
Departments
Information will be available soon.
BI Publications
2025 | |
| |
| |
| |
| |
| |
2024 | |
| |
| |
2023 | |
| |
| |
| |
| |
| |
| |
| |
| |
2022 | |
| |
2021 | |
| |
| |
| |
| |
| |
| |
| |
2020 | |
| |
| |
| |
| |
| |
| |
| |
| |
2019 | |
| |
| |
| |
| |
| |
| |
| |
2018 | |
| |
| |
| |
| |
2017 | |
| |
| |
| |
| |
| |
2016 | |
| |
| |
| |
2015 | |
| |
| |
| |
2014 | |
| |
| |
| |
| |
| |
| |
| |
2013 | |
| |
| |
2012 | |
| |
| |
| |
2011 | |
| |
| |
| |
| |
| |
2010 | |
| |
| |
| |
| |
2009 | |
| |
2008 | |
| |
| |
2007 | |
| |
| |
2006 | |
| |
2005 | |
| |
| |
| |
| |
2004 | |
| |
| |
2003 | |
| |
| |
| |
2002 | |
| |
|
|
| |
2001 | |
| |
| |
| |
|
|
| |
2000 | |
| |
|
|
1999 | |
| |
| |
| |
| |
|
Department of Biological DNA Modification
 | Head |
| Saulius KLIMAŠAUSKAS Dr. Habil., FRSC Distinguished Professor | |
| ORCID; Google Scholar; ResearcherID | |
| phone: +370 5 2234350 fax: +370 5 2234367 e-mail: saulius.klimasauskas (at) bti.vu.lt |
Research Staff PhD students
Giedrius VILKAITIS, Ph.D. Bernadeta MASIULIONYTĖ, M.Sc.
Edita KRIUKIENĖ, Ph.D. Kotryna KVEDERAVIČIŪTĖ, M.Sc.
Vaidotas STANKEVIČIUS, Ph.D. Kotryna SKARDŽIŪTĖ, M.Sc.
Miglė TOMKUVIENĖ, Ph.D. Joris BALČIŪNAS, M.Sc.
Liepa GASIULĖ, Ph.D.
Rasa RAKAUSKAITĖ, Ph.D.
Eglė JAKUBAUSKIENĖ, Ph.D.
Inga PEČIULIENĖ, Ph.D.
Milda MICKUTĖ, Ph.D.
Milda NARMONTĖ, Ph.D.
Janina LIČYTĖ, Ph.D.
Audronė RUKŠĖNAITĖ, M.Sc.
Zdislav STAŠEVSKIJ, M.Sc.
Giedrė URBANAVIČIŪTĖ, M.Sc.

Research Overview
AdoMet-dependent methyltransferases (MTases), which represent more than 3% of the proteins in the cell, catalyze the transfer of the methyl group from S-adenosyl-L-methionine (AdoMet) to N-, C-, O- or S-nucleophiles in DNA, RNA, proteins or small biomolecules.
In DNA, enzymatic methylation of nucleobases serves to expand the information content of the genome in organisms ranging from bacteria to mammals. Postreplicative methylation is accomplished by DNA methyltransferases yielding 5-methylcytosine, N4-methylcytosine or N6-methyladenine (Kweon et al., 2019). Genomic DNA methylation is a key epigenetic regulatory mechanism in high eukaryotes. Aberrant DNA methylation correlates with a number of pediatric syndromes and cancer, or predisposes individuals to various other human diseases. However, research into the epigenetic misregulation and its diagnostics is hampered by the limitations of available analytical techniques.
Targeted covalent labeling of biopolymers
Besides their diverse biological roles, DNA MTases are attractive models to study the structural aspects of DNA-protein interaction. Bacterial enzymes recognize an impressive variety (over 200) of short sequences in DNA. Following detailed mechanistic and structural studies of MTases, we turned to repurposing these enzymes sequence-specific covalent modification of DNA and other biopolymers. Our strategy is based on designing novel synthetic analogues of the natural cofactor AdoMet. We have synthesized a series of model AdoMet analogs with sulfonium-bound extended side chains replacing the methyl group. This novel enabling technology named mTAG (methyltransferase-directed Transfer of Activated Groups) is a convenient and robust technique that is suitable for routine laboratory use. In particular, we demonstrated that propargylic side chains can be efficiently transferred by DNA MTases with high sequence- and base-specificity (Lukinavičius et al., 2007, 2012 and 2013; Masevičius et al., 2016; Tomkuvienė et al., 2016; Tomkuvienė et al., 2019 and 2020) offering many potential applications for genomic (Neely et al., 2010) and epigenomic (see below) studies. Moreover, the newly developed cofactors are suitable for targeted transfer of functional groups or other chemical entities to RNA (Tomkuvienė et al., 2012; Plotnikova et al., 2014; Osipenko et al., 2017; Mickutė et al., 2018 and 2021) using appropriate MTases as catalysts.
In the absence of the S-adenosylmethionine cofactor, bacterial cytosine-5 MTases can catalyze catalyze reversible covalent addition of exogenous aliphatic aldehydes to their target residues in DNA, thus yielding corresponding 5-hydroxyalkylcytosines (Liutkevičiūtė et al., 2009). Moreover, our further studies demonstrated the ability of the MTases to direct condensation of aliphatic thiols and selenols with 5-hydroxymethylcytosine in DNA to yield 5-alkylchalcogenomethyl derivatives (Liutkevičiūtė et al., 2011) or decarboxylation of 5-carboxylcytosines (Liutkevičiūtė et al., 2014) in DNA. These atypical reactions demonstrate a surprizing catalytic versatility of these enzymes and pave new ways for the sequence-specific derivatization and analysis of 5-hydroxymethylcytosine in mammalian DNA (Kriukienė et al., 2012; Gibas et al. 2020; Gordevičius et al., 2020).
Novel approaches to epigenome profiling
Genomic DNA methylation is a prevalent epigenetic modification in mammals, which is brought about by three known DNA cytosine-5 methyltransferases (DNMTs). Although DNA methylation has been extensively investigated, many mechanistic aspects of the DNMT action remain obscure due limitations of current analytical techniques. We therefore aim to develop new experimental approaches to genome-wide profiling of DNA methylation for epigenome studies and improved diagnostics. Our approach is based on selective mTAG labeling and enrichment of unmethylated CpG sites (Kriukienė et al. 2013; Labrie et al., 2016) in the genome followed by analysis of the enriched fractions on tiling microarrays (in collaboration with Prof. Art Petronis, CAMH, Toronto, Canada). Recently, we have advanced DNA methylome profiling by developing a high-resolution economical technique named Tethered Oligonucleotide-Primed sequencing, TOP-seq, which exploits non-homologous priming of the DNA polymerase at covalently tagged CpG or hmCpG sites to directly produce adjoining regions for their sequencing and precise genomic mapping (Staševskij et al., 2017; Gibas et al. 2020; Ličytė et al. 2020; Gordevičius et al., 2020).
Single-cell temporal tracking of epigenetic DNA marks (EpiTrack) ![]()
Our ERC-supported studies (Single-cell temporal tracking of epigenetic DNA marks, EpiTrack) aimed to gain in-depth understanding of how the genomic methylation patterns are established and how they govern cell plasticity and variability during differentiation and development. These questions were addressed by precise determination of where and when methylation marks are deposited by the individual DNMTs, and how these methylation marks affect gene expression. To achieve this goal, we use metabolic engineering of mouse cells to permit SAM analog-based chemical pulse-tagging of their methylation sites in vivo to unveil, with unprecedented detail, the dynamics and variability of DNA methylation during differentiation of mouse embryonic cells to somatic lineages.
Methylation of small non-coding RNA
MicroRNAs and siRNAs are small non-coding double-stranded RNA molecules that control gene activity in a homology-dependent manner - a process named RNA interference. Since their discovery in 1993, numerous microRNAs have been identified and recognized as important regulators of gene expression in both plants and animals. Many microRNAs have well-defined developmental and tissue-specific expression pattern, but a great number of microRNAs and their roles are still unknown.
HEN1 methyltransferases from plants and animals catalyze the transfer methyl groups from AdoMet onto the 2'OH group of the 3'-terminal nucleotide of small RNAs, like miRNA, siRNA/siRNA or piRNA. The methylation is imperative in the biogenesis of microRNA in plants and piRNA in animals. A number of chemo-enzymatic approaches have been developed in our laboratory for examining and exploiting the unique properties of the HEN1 methyltransferases (Plotnikova et al., 2013; Baranauskė et al., 2015; Osipenko et al., 2017; Mickutė et al., 2018, 2021).
Recent publications
V. Stankevičius, L. Gasiulė, G. Vilkaitis, S. Klimašauskas
Editorial: Selective chemical tracking of DNA methylomes in live cells.
Epigenomics, 2025, 17(9): 575-577. PDF
L. Gasiulė, V. Stankevičius, K. Kvederavičiūtė, J. M. Rimšelis, V. Klimkevičius, G. Petraitytė, A. Rukšėnaitė, V. Masevičius, S. Klimašauskas
Engineered methionine adenosyltransferase cascades for metabolic labeling of individual DNA methylomes in live cells.
J. Am. Chem. Soc.,2024, 146(27): 18722-18729.
E. Kriukienė, M. Tomkuvienė, S. Klimašauskas
5-Hydroxymethylcytosine: the many faces of the sixth base of mammalian DNA.
Chem. Soc. Rev., 2024, 53: 2264–2283.
K. Skardžiūtė, K. Kvederavičiūtė, I. Pečiulienė, M. Narmontė, P. Gibas, J. Ličytė, S. Klimašauskas, E. Kriukienė
One-pot trimodal mapping of unmethylated, hydroxymethylated and open chromatin sites unveils distinctive 5hmC roles at dynamic chromatin loci.
Cell Chem. Biol., 2024, 31(3): 607-621.e9.
G. Vilkaitis, V. Masevičius, E. Kriukienė, S. Klimašauskas
Chemical Expansion of the Methyltransferase Reaction: Tools for DNA Labeling and Epigenome Analysis.
Acc. Chem. Res., 2023, 56: 3188-3197.
M. Mickutė, R. Krasauskas, K. Kvederavičiūtė, G. Tupikaitė, A. Osipenko, A. Kaupinis, M. Jazdauskaitė, R. Mineikaitė, M. Valius, V. Masevičius, G. Vilkaitis
Interplay between bacterial 5′-NAD-RNA decapping hydrolase NudC and DEAD-box RNA helicase CsdA in stress responses.
mSystems, 2023, 8: e00718-23.
M. Malikėnas, V. Masevičius, S. Klimašauskas
Synthesis of S-adenosyl-L-methionine analogs with extended transferable groups for methyltransferase-directed labeling of DNA and RNA.
Curr. Protoc., 2023, 3: e799.
M. Tomkuvienė, M. Meier, D. Ikasalaitė, J. Wildenauer, V. Kairys, S. Klimašauskas, L. Manelytė
Enhanced nucleosome assembly at CpG sites containing an extended 5-methylcytosine analogue.
Nucleic Acids Res., 2022, 50(11): 6549–6561.
M.J. Peña-Gómez, P. Moreno-Gordillo, M. Narmontė, C.B. García-Calderón, A. Rukšėnaitė, S. Klimašauskas, I.V. Rosado
FANCD2 maintains replication fork stability during misincorporation of the DNA demethylation products 5-hydroxymethyl-2'-deoxycytidine and 5-hydroxymethyl-2'-deoxyuridine.
Cell Death Dis., 2022, 13(5): 503.
V. Stankevičius, P. Gibas, B. Masiulionytė, L. Gasiulė, V. Masevičius, S. Klimašauskas, G. Vilkaitis
Selective chemical tracking of Dnmt1 catalytic activity in live cells.
Mol. Cell, 2022, 82(5): 1053-1065. --Meet the author interview.
J. Ličytė, K. Kvederavičiūtė, A. Rukšėnaitė, E. Godliauskaitė, P. Gibas, V. Tomkutė, G. Petraitytė, V. Masevičius, S. Klimašauskas, E. Kriukienė
Distribution and regulatory roles of oxidized 5-methylcytosines in DNA and RNA of the Basidiomycete fungi Laccaria bicolor and Coprinopsis cinerea.
Open Biol., 2022, 12(3): 210302.
M. Tomkuvienė, E. Kriukienė, S. Klimašauskas
DNA labeling using DNA methyltransferases.
Adv. Exp. Med. Biol., 2022, 1389: 535–562.
M. Narmontė, P. Gibas, K. Daniūnaitė, J. Gordevičius, E. Kriukienė
Multi-omics analysis of neuroblastoma cells reveals a diversity of malignant transformations.
Front. Cell Dev. Biol., 2021, 9: 727353.
M. Mickutė, K. Kvederavičiūtė, A. Osipenko, R. Mineikaitė, S. Klimašauskas, G. Vilkaitis
Methyltransferase-directed orthogonal tagging and sequencing of miRNAs and bacterial small RNAs.
BMC Biology, 2021, 19: 129.
A.N. Tesfahun, M. Alexeeva, M. Tomkuvienė, A. Arshad, P. Guragain, A. Klungland, S. Klimašauskas, P. Ruoff, S. Bjelland
Alleviation of C-C Mismatches in DNA by the Escherichia coli Fpg Protein.
Front. Microbiol., 2021, 12: 608839.
R. Rakauskaitė, G. Urbanavičiūtė, M. Simanavičius, A. Žvirblienė, S. Klimašauskas
Selective immunocapture and light-controlled traceless release of transiently caged proteins.
STAR Protoc., 2021, 2(2): 100455.
R. Rakauskaitė, G. Urbanavičiūtė, M. Simanavičius, R. Lasickienė, A. Vaitiekaitė, G. Petraitytė, V. Masevičius, A. Žvirblienė, S. Klimašauskas
Photocage-Selective Capture and Light-Controlled Release of Target Proteins.
iScience, 2020, 23(12): 101833.
M. Tomkuvienė, D. Ikasalaitė, A. Slyvka, A. Rukšėnaitė, M. Ravichandran, T. P. Jurkowski, M. Bochtler, S. Klimašauskas
Enzymatic hydroxylation and excision of extended 5-methylcytosine analogues.
J. Mol. Biol., 2020, 423(23): 6157-6167.
J. Gordevičius, M. Narmontė, P. Gibas, K. Kvederavičiūtė, V. Tomkutė, P. Paluoja, K. Krjutškov, A. Salumets, E. Kriukienė
Identification of fetal unmodified and 5-hydroxymethylated CG sites in maternal cell-free DNA for non-invasive prenatal testing.
Clin. Epigen., 2020, 12: 153.
J. Ličytė, P. Gibas, K. Skardžiūtė, V. Stankevičius, A. Rukšėnaitė, E. Kriukienė
A Bisulfite-free Approach for Base-Resolution Analysis of Genomic 5-Carboxylcytosine.
Cell Rep., 2020, 32(11): 108155.
P. Gibas, M. Narmontė, Z. Staševskij, J. Gordevičius, S. Klimašauskas, E. Kriukienė
Precise genomic mapping of 5-hydroxymethylcytosine via covalent tether-directed sequencing.
PLOS Biol., 2020, 18(4): e3000684.
S. Gasiulė, N. Dreize, A. Kaupinis, R. Ražanskas, L. Čiupas, V. Stankevičius, Ž. Kapustina, A. Laurinavičius, M. Valius, G. Vilkaitis
Molecular Insights into miRNA-Driven Resistance to 5-Fluorouracil and Oxaliplatin Chemotherapy: miR-23b Modulates the Epithelial–Mesenchymal Transition of Colorectal Cancer Cells.
J. Clin. Med., 2019, 8(12): 2115.
S. Gasiulė, V. Stankevičius, V. Patamsytė, R. Ražanskas, G. Žukovas, Ž. Kapustina, D. Žaliaduonytė, R. Benetis, V. Lesauskaitė, G. Vilkaitis
Tissue-Specific miRNAs Regulate the Development of Thoracic Aortic Aneurysm: The Emerging Role of KLF4 Network.
J. Clin. Med., 2019, 8(10): 1609.
S.-M. Kweon, Y. Chen, E. Moon, K. Kvederavičiūtė, S. Klimašauskas, D.E. Feldman
An adversarial DNA N6-methyladenine-sensor network preserves polycomb silencing.
Mol. Cell, 2019, 74(6): 1138-1147.e6.
M. Tomkuvienė, M. Mickutė, G. Vilkaitis, S. Klimašauskas
Repurposing enzymatic transferase reactions for targeted labeling and analysis of DNA and RNA.
Curr. Opin. Biotechnol., 2019, 55: 114-123.
K. Daniūnaitė, S. Jarmalaitė, E. Kriukienė
Epigenomic technologies for diciphering circulating tumor DNA.
Curr. Opin. Biotechnol., 2019, 55: 23-29.
M. Mickutė, M. Nainytė, L. Vasiliauskaitė. A. Plotnikova, V. Masevičius, S. Klimašauskas, G. Vilkaitis
Animal Hen1 2′-O-methyltransferases as tools for 3′-terminal functionalization and labelling of single-stranded RNAs.
Nucleic Acids Res., 2018, 46: e104.
M. Alexeeva, P. Guragain, A.N. Tesfahun, M. Tomkuvienė, A. Arshad, R. Gerasimaitė, A. Rukšėnaitė, G. Urbanavičiūtė, M. Bjørås, J.K. Laerdahl, A. Klungland, S. Klimašauskas, S. Bjelland
Excision of the double methylated base N4,5-dimethylcytosine from DNA by Escherichia coli Nei and Fpg proteins.
Phil. Trans. R. Soc. B, 2018, 373(1748): 20170337.
M. Tomkuvienė, J. Ličytė, I. Olendraitė, Z. Liutkevičiūtė, B. Clouet-d'Orval, S. Klimašauskas
Archaeal fibrillarin-Nop5 heterodimer 2'-O-methylates RNA independently of the C/D guide RNP particle.
RNA, 2017, 23(9): 1329-1337.
A. Osipenko, A. Plotnikova, M. Nainytė, V. Masevičius, S. Klimašauskas, G. Vilkaitis
Oligonucleotide-addressed covalent 3’-terminal derivatization of small RNA strands for enrichment and visualization.
Angew. Chem. Int. Ed., 2017, 56(23): 6507–6510.
Z. Staševskij, P. Gibas, J. Gordevičius, E. Kriukienė, S. Klimašauskas
Tethered Oligonucleotide-Primed sequencing, TOP-seq: a high resolution economical approach for DNA epigenome profiling.
Mol. Cell, 2017, 65(3): 554–564.
M. Tomkuvienė, E. Kriukienė, S. Klimašauskas
DNA labeling using DNA methyltransferases.
Adv. Exp. Med. Biol., 2016, 945: 511-535.
V. Labrie, O. J. Buske, E. Oh, R. Jeremian, C. Ptak, G. Gasiūnas, A. Maleckas, R. Petereit, A. Žvirbliene, K. Adamonis, E. Kriukienė, K. Koncevičius, J. Gordevičius, A. Nair, A. Zhang, S. Ebrahimi, G. Oh, V. Šikšnys, L. Kupčinskas, M. Brudno, A. Petronis
Lactase nonpersistence is directed by DNA-variation-dependent epigenetic aging.
Nature Struct. Mol. Biol. 2016, 23(6): 566-573.
V. Myrianthopoulos, P. F. Cartron, Z. Liutkevičiūtė, S. Klimašauskas, D. Matulis, C. Bronner, N. Martinet, E. Mikros
Tandem virtual screening targeting the SRA domain of UHRF1 identifies a novel chemical tool modulating DNA methylation.
Eur. J. Med. Chem., 2016, 114: 390–396.
V. Masevičius, M. Nainytė, S. Klimašauskas
Synthesis of S-adenosyl-L-methionine analogs with extended transferable groups for methyltransferase-directed labeling of DNA and RNA.
Curr. Protoc. Nucleic Acid Chem., 2016, 64: 1.36.1-1.36.13.
R. Rakauskaitė, G. Urbanavičiūtė, A. Rukšėnaitė, Z. Liutkevičiūtė, R. Juškėnas, V. Masevičius, S. Klimašauskas
Biosynthetic selenoproteins with geneticallyencoded photocaged selenocysteines.
Chem. Commun., 2015, 51(39): 8245-8248.
S. Baranauskė, M. Mickutė, A. Plotnikova, A. Finke, Č. Venclovas, S. Klimašauskas, G. Vilkaitis
Functional mapping of the plant small RNAmethyltransferase: HEN1 physically interacts with HYL1 and DICER-LIKE 1 proteins.
Nucleic Acids Res., 2015, 43(5): 2802-2812.
A. Plotnikova, A. Osipenko, V. Masevičius, G. Vilkaitis, S. Klimašauskas
Selective covalent labeling of miRNA and siRNA duplexes using HEN1 methyltransferase.
J. Am. Chem. Soc., 2014, 136(39): 13550–13553.
Z. Liutkevičiūtė, E. Kriukienė, J. Ličytė, M. Rudytė, G. Urbanavičiūtė, S. Klimašauskas
Direct decarboxylation of 5-carboxylcytosine by DNA C5-methyltransferases.
J. Am. Chem. Soc., 2014, 136(16): 5884−5887.
Department of Eukaryote Gene Engineering

Head: Dr. Rasa Burneikienė-Petraitytė
Tel. +370-5-2234421
Fax. +370-5-2687009
E-mail: rasa.burneikiene @ bti.vu.lt
Habil. Dr. Alma Gedvilaitė’s group activities:
The activities of the research group addresses studies related to efficient production of recombinant viral proteins in yeast cells by improvement of expression systems and yeast genetic background. Despite that yeast Saccharomyces cerevisiae frequently serves as a reasonable host for heterologous protein expression, in many instances, it is far from optimal. Our aim is to understand and balance processes in yeast trigged by synthesis of recombinant proteins, identify factors necessary for efficient recombinant protein expression by overexpression or deletion of target genes. In an attempt to elucidate the requirement of factors for the biosynthesis of recombinant viral and human proteins we use proteomics, yeast mutant and gene collection studies. Very important part of group study is related to improvement of yeast protein secretion capacities by identification and investigation of yeast genes conferring supersecretion phenotypes.
The group is also interested in search and characterization of new polyomaviruses, studies of virus-like particles (VLPs) formation, and protein engineering based on construction of chimeric VLPs harboring foreign epitopes. We employ hamster polyomavirus VLPs as carriers for short and non-immunogenic protein sequences with are presented on VP1 VLPs to increase their immunogenicity. For presentation of large and complex protein molecules (as single chain antibody molecules) we use VP1/VP2 VLPs.
We have a wide network of international collaboration and long-lasting experience in conducting collaborative projects with industrial partners.
Current project:
Project of Research Council of Lithuania "Investigation of Kluyveromyces lactis mutations conferring enhanced secretion phenotype and generation of yeast strains for supersecretion of recombinant proteins", 2017-2021, project leader A.Gedvilaite.
Results:
Identified four new common and bank voles, yellow-necked mouse and shrews polyomaviruses.
Large collection of more than 40 different virus like particles (VLPs) derived from various polyomavirus (all 13 human polyomavirus, rodents, avian and other host polyomavirus) VP1 proteins and papillomavirus L1 proteins (HPV6, 11, 16, 18, 31, 33), porcine circovirus type 2 capsid proteins.
Yeast expression systems for construction and production of hamster polyomavirus VP1 and VP1/VP2 VLPs as carriers harboring non immunogenic peptides or complex molecules of interest.
Genetic engineered yeast strain collection for recombinant protein production.
2003 – Lithuanian Science Prize in the Biomedicine Sciences area.
Patent: Zvirblienė A., Gedvilaitė A., Ulrich, R., Sasnauskas. Process for the production of monoclonal antibodies using chimeric VLPs. US Patent No.:US7,919,314 B2. Apr. 5, 2011.
Selected publications:
Gedvilaite A, Tryland M, Ulrich RG, Schneider J, Kurmauskaite V, Moens U, Preugschas H, Calvignac-Spencer S, Ehlers B. Novel polyomaviruses in shrews (Soricidae) with close similarity to human polyomavirus 12. J Gen Virol. 2017. doi: 10.1099/jgv.0.000948.
Zaveckas M, Goda K, Ziogiene D, Gedvilaite A. Purification of recombinant trichodysplasia spinulosa-associated polyomavirus VP1-derived virus-like particles using chromatographic techniques. J Chromatogr B Analyt Technol Biomed Life Sci. 2018 May 9;1090:7-13. doi: 10.1016/j.jchromb.2018.05.007.
Valaviciute M, Norkiene M, Goda K, Slibinskas R, Gedvilaite A. Survey of molecular chaperone requirement for the biosynthesis of hamster polyomavirus VP1 protein in Saccharomyces cerevisiae. Arch Virol. 2016. 161(7), 1807-1819. DOI 10.1007/s00705-016-2846-3
Nainys J, Timinskas A, Schneider J, Ulrich RG, Gedvilaite A. Identification of Two Novel Members of the Tentative Genus Wukipolyomavirus in Wild Rodents. PLoS One. 2015 Oct 16;10(10):e0140916. doi: 10.1371/journal.pone.0140916. eCollection 2015.
Pleckaityte M, Bremer CM, Gedvilaite A, Kucinskaite-Kodze I, Glebe D, Zvirbliene A. Construction of polyomavirus-derived pseudotype virus-like particles displaying a functionally active neutralizing antibody against hepatitis B virus surface antigen. BMC Biotechnology. 2015, 15:85. DOI 10.1186/s12896-015-0203-3
Norkiene M, Stonyte J, Ziogiene D, Mazeike E, Sasnauskas K, Gedvilaite A. Production of recombinant VP1-derived virus-like particles from novel human polyomaviruses in yeast. BMC Biotechnology. 2015, 15:68. DOI: 10.1186/s12896-015-0187-z
Zaveckas M., Snipaitis S., Pesliakas H. Nainys J. Gedvilaite A. Purification of recombinant virus-like particles of porcine circovirustype 2 capsid protein using ion-exchange monolith chromatography. J Chromatogr B. 2015, v. 991, p.21–28. doi:10.1016/j.jchromb.2015.04.004
Nainys J, Lasickiene R, Petraityte-Burneikiene R, Dabrisius J, Lelesius R, Sereika V, Zvirbliene A, Sasnauskas K, Gedvilaite A. Generation in yeast of recombinant virus-like particles of porcine circovirus type 2 capsid protein and their use for a serologic assay and development of monoclonal antibodies. BMC Biotechnology 2014 14:100. doi:10.1186/s12896-014-0100-1 847
Pleckaityte M, Zvirbliene A, Sezaite I, Gedvilaite A. Production in yeast of pseudotype virus-like particles harboring functionally active antibody fragments neutralizing the cytolytic activity of vaginolysin. Microb Cell Fact. 2011. 10:109.
Gedvilaite A., Dorn D.C., Sasnauskas K., Pecher G., Bulavaite A., Lawatscheck R., Staniulis J., Dalianis T., Ramqvist T., Schönrich G., Raftery MJ., Ulrich R. Virus-like particles derived from major capsid protein VP1 of different polyomaviruses differ in their ability to induce maturation in human dendritic cells. Virology, 2006, v. 354 (2), 252-260.
Gedvilaite A., Frommel C., Sasnauskas K., Burkhard M., Ozel M., Behrsing O., Staniulis J., Jandrig B., Scherneck S., Ulrich R. Formation of Immunogenic Virus-like Particles by Inserting Epitopes into Surface-Exposed regions of Hamster Major Capsid Protein // Virology. 2000, v. 273, n. 1, p. 21-35.
Group of Drs. Rimantas Slibinskas and Evaldas Čiplys
Research:
Our group is studying processes of recombinant protein synthesis in yeast expression systems by using various methods including proteomic analysis. Research is mainly focused on foreign protein processing in cell secretion pathway and related stress responses. Essential molecular components for proper functioning of secretory pathway and for efficient secretion of recombinant proteins are chaperones located in endoplasmic reticulum (ER). We found that human ER chaperones are correctly processed and efficiently produced in the active form in yeast cells suggesting compatibility of human and yeast cellular machinery for synthesis and processing of these specific proteins. Moreover, yeast cells secreted large amounts of human ER chaperones BiP/GRP78, calreticulin and ERp57 under conditions of overexpression. It is known that translocation of these proteins to the cell surface or their secretion outside the cells in human tissues is related to various pathological conditions with connection to a number of human diseases. Therefore, it makes yeast expression systems an excellent model for studying molecular mechanisms of the secretion of human ER chaperones. Furthermore, high quality and widely affordable recombinant human ER chaperone proteins are needed for further research and studies of the potential application of these proteins. For this purpose we also develop technologies for the production of human ER chaperones and some of their mutants in yeast.
Current project:
Research and development project, partially supported by the EU structural funds, „Development of technology for production of native recombinant human calreticulin in yeast“ (2016-2019), project leaders R. Slibinskas and E. Čiplys.
Selected publications:
Overexpression of human virus surface glycoprotein precursors induces cytosolic unfolded protein response in Saccharomyces cerevisiae. Ciplys E, Samuel D, Juozapaitis M, Sasnauskas K, Slibinskas R.Microb Cell Fact. 2011 May 19;10:37. doi: 10.1186/1475-2859-10-37.
Overexpression of human calnexin in yeast improves measles surface glycoprotein solubility. Ciplys E, Sasnauskas K, Slibinskas R.FEMS Yeast Res. 2011 Sep;11(6):514-23. doi: 10.1111/j.1567-1364.2011.00742.x. Epub 2011 Jun 27.
Native signal peptide of human ERp57 disulfide isomerase mediates secretion of active native recombinant ERp57 protein in yeast Saccharomyces cerevisiae.Čiplys E, Žitkus E, Slibinskas R. Protein Expr Purif. 2013 Jun;89(2):131-5. doi: 10.1016/j.pep.2013.03.003. Epub 2013 Mar 23.
Comparison of first dimension IPG and NEPHGE techniques in two-dimensional gel electrophoresis experiment with cytosolic unfolded protein response in Saccharomyces cerevisiae. Slibinskas R, Ražanskas R, Zinkevičiūtė R, Čiplys E. Proteome Sci. 2013 Jul 27;11(1):36. doi: 10.1186/1477-5956-11-36.
Generation of human ER chaperone BiP in yeast Saccharomyces cerevisiae. Čiplys E, Aučynaitė A, Slibinskas R. Microb Cell Fact. 2014 Feb 11;13:22.doi: 10.1186/1475-2859-13-22.
Calreticulin as cancer treatment adjuvant: combination with photodynamic therapy and photodynamic therapy-generated vaccines. Korbelik M, Banáth J, Saw KM, Zhang W, Čiplys E. Front Oncol. 2015 Feb 3;5:15. doi: 10.3389/fonc.2015.00015.
Heat shock at higher cell densities improves measles hemagglutinin translocation and human GRP78/BiP secretion in Saccharomyces cerevisiae. Zinkevičiūtė R, Bakūnaitė E, Čiplys E, Ražanskas R, Raškevičiūtė J, Slibinskas R. N Biotechnol. 2015 Dec 25;32(6):690-700. doi: 10.1016/j.nbt.2015.04.001. Epub 2015 Apr 20.
High-level secretion of native recombinant human calreticulin in yeast. Čiplys E, Žitkus E, Gold LI, Daubriac J, Pavlides SC, Højrup P, Houen G, Wang WA, Michalak M, Slibinskas R. Microb Cell Fact. 2015 Oct 15;14:165.doi: 10.1186/s12934-015-0356-8.
Mapping the Ca(2+) induced structural change in calreticulin. Boelt SG, Norn C, Rasmussen MI, André I, Čiplys E, Slibinskas R, Houen G, Højrup P. J Proteomics. 2016 Jun 16;142:138-48. doi: 10.1016/j.jprot.2016.05.015. Epub 2016 May 16.
Survey of molecular chaperone requirement for the biosynthesis of hamster polyomavirus VP1 protein in Saccharomyces cerevisiae. Valaviciute M, Norkiene M, Goda K, Slibinskas R, Gedvilaite A. Arch Virol. 2016 Jul;161(7):1807-19. doi: 10.1007/s00705-016-2846-3.
Group of Dr. Rasa Petraitytė-Burneikienė
Analysis, synthesis of viral proteins in yeast and their practical application is the main research field of our group. We are focusing on aspects related to the production of recombinant proteins in yeast expression systems, the development and optimization of expression systems dedicated to the production of recombinant proteins as virus-like particles. The core research techniques used in the department are the methods of molecular biology and biotechnology, leading to obtain recombinant plasmids, genetically modified bacteria and yeast, and purification of proteins. We have already in our disposition powerful diagnostic tools as yeast-expressed recombinant proteins for diagnostics of human hantaviruses, porcine parvovirus, human bocaviruses 1-4, human metapneumovirus, human parvovirus 4, parainfluenza viruses 2 and 4, hepatitis E and bovine Schmallenberg virus. Yeast expressed recombinant proteins are applied in the tests for detection of virus-specific antibodies in human serum and oral fluid samples.
Current projects:
1. National programme “Healthy Aging”, Research Council of Lithuania. Investigation of synthesis regulation of proteins associated with Alzheimer disease development (No. SEN-05/2015). K. Sasnauskas. 2015–2018.
2. Joint Lithuanian–Latvian–Chinese (Taiwanese) Tripartite Cooperation Programme. Research Council of Lithuania. Studying of human parvovirus B19, bocavirus and parvovirus 4 involvement in inflammatory neurological diseases using interdisciplinary approach (No. TAP LLT-3/2017). R. Petraitytė-Burneikienė. 2017-2019.
3. LSC project for activity according action 2015-LMT-K-718 “Scientific research for advanced investigators“ (Smart specialization) , No. 01.2.2-LMT-K-718-01-0008 ,,New technologies for construction of recombinant allergens“, G. Žvirblis, 2018-2021.
Collaboration
Friedrich-Loeffler-Institut Bundesforschungsinstitut für Tiergesundheit, Federal Research
Institute for Animal Health OIE Collaborating Centre for Zoonoses in Europe (Germany)
Department of Virology, University of Freiburg (Germany)
A. Kirchenstein Institute of Microbiology and Virology, Riga Stradins University (Latvia)
Department of Medical Research, Mackay Memorial Hospital, (Taiwan)
Selected publications:
1. Bulavaitė A, Lasickienė R, Vaitiekaitė A, Sasnauskas K, Žvirblienė A. (2016). Synthesis of human parainfluenza virus 2 nucleocapsid protein in yeast as nucleocapsid-like particles and investigation of its antigenic structure. Appl Microbiol Biotechnol. 100(10):4523-34.
2. Chandy, S., Ulrich, R.G., Schlegel, M., Petraityte, R., Sasnauskas, K., Prakash, D.J., Balraj, V., Abraham, P., Sridharan, G. (2013). Hantavirus Infection among Wild Small Mammals in Vellore, South India. Zoonoses Public Health. 60(5):336-40.
3. Dargevicius, A., Petraityte, R., Sribikiene, B., Sileikiene, E., Razukeviciene, L., Ziginskiene, E., Vorobjoviene, R., Razanskiene, A., Sasnauskas, K., Bumblyte, I.A., Kuzminskis, V.I. (2007). Prevalence of antibodies to hantavirus among hemodialysis patients with end-stage renal failure in Kaunas and its district. Medicina (Kaunas). 43 Suppl. 1:72-6.
4. Emuzyte, R., Firantiene, R., Petraityte, R., Sasnauskas, K. (2009). Human rhinoviruses, allergy, and asthma: a clinical approach. Medicina (Kaunas). 45(11):839-47.
5. Essbauer, S.S., Schmidt-Chanasit, J., Madeja, E.L., Wegener, W., Friedrich, R., Petraityte, R., Sasnauskas, K., Jacob, J., Koch, J., Dobler, G., Conraths, F.J., Pfeffer, M., Pitra, C., and Ulrich, R.G. (2007). Nephropathia epidemica in metropolitan area, Germany. Emerg. Infect. Dis. 13(8):1271-1273.
6. Gedvilaitė A., Dorn D.C., Sasnauskas K., Pecher G., Bulavaitė A., Lawatscheck R., Staniulis J., Dalianis T., Ramqvist T., Schönrich G., Raftery M.J. and Ulrich,R. Virus-like particles derived from major capsid protein VP1 of different polyomaviruses differ in their entry pathways and ability to induce maturation in human dendritic cells. Virology // ISSN: 0042-6822, Academic Press, New York, 2006,354 (2), 252-260.
7. Kucinskaite-Kodze I, Pleckaityte M, Bremer CM, Seiz PL, Zilnyte M, Bulavaite A, Mickiene G, Zvirblis G, Sasnauskas K, Glebe D, Zvirbliene A. (2016). New broadly reactive neutralizing antibodies against hepatitis B virus surface antigen. Virus Res. 211:209-21.
8. Kucinskaite-Kodze, I., Petraityte-Burneikiene, R., Zvirbliene, A., Hjelle, B., Medina, R.A., Gedvilaite, A., Razanskiene, A., Schmidt-Chanasit, J., Mertens, M., Padula, P., Sasnauskas, K., Ulrich, R.G. (2011). Characterization of monoclonal antibodies against hantavirus nucleocapsid protein and their use for immunohistochemistry on rodent and human samples. Arch Virol. 156(3):443-56
9. Lazutka, J., Spakova, A., Sereika, V., Lelesius, R., Sasnauskas, K., Petraityte-Burneikiene, R. (2015). Saliva as an alternative specimen for detection of Schmallenberg virus-specific antibodies in bovines. BMC Veterinary Research. 11(1):237
10. Lazutka, J., Zvirbliene, A., Dalgediene, I., Petraityte-Burneikiene, R., Spakova, A., Sereika, V., Lelesius, R., Wernike, K., Beer, M., Sasnauskas, K. (2014). Generation of Recombinant Schmallenberg Virus Nucleocapsid Protein in Yeast and Development of Virus-Specific Monoclonal Antibodies. Journal of Immunology Research. 2014:160316.
11. Mertens, M., Hofmann, J., Petraityte-Burneikiene, R., Ziller, M., Sasnauskas, K., Friedrich, R., Niederstrasser, O., Krüger, D.H., Groschup, M.H., Petri, E., Werdermann, S., Ulrich, R.G. (2011). Seroprevalence study in forestry workers of a non-endemic region in eastern Germany reveals infections by Tula and Dobrava-Belgrade hantaviruses. Med. Microbiol. Immunol. 200(4):263-8.
12. Mertens, M., Kindler, E., Emmerich, P., Esser, J., Wagner-Wiening, C., Wölfel, R., Petraityte-Burneikiene, R., Schmidt-Chanasit, J., Zvirbliene, A., Groschup, M.H., Dobler, G., Pfeffer, M., Heckel, G., Ulrich, R.G., Essbauer, S.S. (2011). Phylogenetic analysis of Puumala virus subtype Bavaria, characterization and diagnostic use of its recombinant nucleocapsid protein. Virus Genes. 43(2):177-91.
13. Nainys, J., Lasickiene, R., Petraityte-Burneikiene, R., Dabrisius, J., Lelesius, R., Sereika, V., Zvirbliene, A., Sasnauskas, K., Gedvilaite, A. (2014). Generation in yeast of recombinant virus-like particles of porcine circovirus type 2 capsid protein and their use for a serologic assay and development of monoclonal antibodies. BMC Biotechnology. 9;14(1):100.
14. Petraitytė, R., Jin, L., Hunjan, R., Ražanskienė, A., Žvirblienė, A., Sasnauskas, K. (2007). Use of Saccharomyces cerevisiae-expressed recombinant nucleocapsid protein to detect Hantaan virus-specific immunoglobulinG (IgG) and IgM in oral fluid. Clin. Vaccine Immunol. 14(12):1603-1608.
15. Petraityte, R., Tamosiunas, P.L., Juozapaitis, M., Zvirbliene, A., Sasnauskas, K., Shiell, B., Russell, G., Bingham, J., Michalski, W.P. (2009). Generation of Tioman virus nucleocapsid-like particles in yeast Saccharomyces cerevisiae. Virus Res. 145(1):92-6.
16. Petraityte, R., Yang, H., Hunjan, R., Razanskiene, A., Dhanilall, P., Ulrich, R.G., Sasnauskas, K., Jin, L. (2008). Development and evaluation of serological assays for detection of Hantaanvirus-specific antibodies in human sera using yeast-expressed nucleocapsid protein. J. Virol. Methods. 148(1-2):89-95.
17. Petraitytė-Burneikienė, R., Nalivaiko, K., Lasickienė, R., Firantienė, R., Emužytė, R., Sasnauskas, K., Zvirblienė, A. (2011). Generation of recombinant metapneumovirus nucleocapsid protein as nucleocapsid-like particles and development of virus-specific monoclonal antibodies. Virus Res. 161(2):131-9.
18. Schlegel, M., Tegshduuren, E., Yoshimatsu, K., Petraityte, R., Sasnauskas, K., Hammerschmidt, B., Friedrich, R., Mertens, M., Groschup, M.H., Arai, S., Endo, R., Shimizu, K., Koma, T., Yasuda, S., Ishihara, C., Ulrich, R.G., Arikawa, J., Köllner, B. (2012). Novel serological tools for detection of Thottapalayam virus, a Soricomorpha-borne hantavirus. Arch. Virol. 157(11):2179-87.
19. Schmidt, J., Meisel, H., Capria, S.G., Petraityte, R., Lundkvist, A., Hjelle, B., Vial, P.A., Padula, P., Kruger, D.H., Ulrich, R. (2006). Serological assays for the detection of human andes hantavirus infections based on its yeast-expressed nucleocapsid protein. Intervirology. 49(3):173-84.
20. Schmidt-Chanasit, J., Essbauer, S., Petraityte, R., Yoshimatsu, K., Tackmann, K., Conraths, F.J., Sasnauskas, K., Arikawa, J., Thomas, A., Pfeffer, M., Scharninghausen, J.J., Splettstoesser, W., Wenk, M., Heckel, G., Ulrich R.G. (2010). Extensive host sharing of central European Tula virus. J Virol. 84(1):459-74.
21. Skrastina, D., Bulavaite, A. Sominskaja, I., Kovalevska, L., Ose, V., Priede, D., Pumpens, P., Sasnauskas, K. Immunological behaviour of immunodominant hepatitis B virus envelope exposed on the surface of two different virus-like particle carriers. Vaccine, 2008, 26, 1972-1981.
22. Skrastina, D., Petrovskis, I., Petraityte, R., Sominskaya, I., Ose, V., Lieknina, I., Bogans, J., Sasnauskas, K., Pumpens, P. (2013). Chimeric derivatives of hepatitis B virus core particles carrying major epitopes of the rubella virus E1 glycoprotein. Clin. Vaccine Immunol. 20(11):1719-28.
23. Tamošiūnas, P. L., Petraitytė-Burneikienė, R., Lasickienė, R., Akatov, A., Kundrotas, G., Sereika, V., Lelešius, R., Zvirblienė, A., Sasnauskas, K. (2014). Generation of Recombinant Porcine Parvovirus Virus-Like Particles in Saccharomyces cerevisiae and Development of Virus-Specific Monoclonal Antibodies. Journal of Immunology Research. 2014:573531.
24. Tamošiūnas, P. L., Simutis, K., Kodzė, I., Firantienė, R., Emužytė, R., Petraitytė-Burneikienė, R., Zvirblienė, A., Sasnauskas, K. (2013). Production of Human Parvovirus 4 VP2 Virus-Like Particles in Yeast and Their Evaluation as an Antigen for Detection of Virus-Specific Antibodies in Human Serum. Intervirology. 56(5):271-7.
25. Tamošiūnas, P.L., Petraitytė-Burneikienė, R., Bulavaitė, A., Marcinkevičiūtė, K., Simutis, K., Lasickienė, R., Firantienė, R., Ėmužytė, R., Žvirblienė, A., Sasnauskas, K. (2016). Yeast-generated virus-like particles as antigens for detection of human bocavirus 1-4 specific antibodies in human serum. Applied Microbiology and Biotechnology. 100(11):4935-46.
26. Zvirbliene, A., Kucinskaite-Kodze, I., Razanskiene, A., Petraityte-Burneikiene, R., Klempa, B., Ulrich, R.G., Gedvilaite, A. (2014). The Use of Chimeric Virus-like Particles Harbouring a Segment of Hantavirus Gc Glycoprotein to Generate a Broadly-Reactive Hantavirus-Specific Monoclonal Antibody. Viruses. 6, 640-660.
Department of Immunology and Cell Biology
Monoclonal and recombinant antibodies are widely used in biotechnology, medicine and biomedical science. Monoclonal antibodies produced using traditional hybridoma-based technologies are valuable research tools and clinical diagnostic reagents. Recombinant antibodies generated by gene engineering approaches are increasingly being used as therapeutic agents for treatment of cancer, autoimmune and infectious diseases. So, there is strong need for novel well-characterized antibodies with desired specificities and other characteristics.
Our team
Our team has a strong expertise in development and characterization of monoclonal and recombinant antibodies. We have generated more than 500 monoclonal antibodies against different targets: viral antigens, bacterial virulence factors, cellular proteins, cytokines, hormones. The largest antibody collection is generated against viral antigens, including measles, mumps, human parainfluenza viruses, henipaviruses, hantaviruses, parvoviruses, human bocaviruses, hepatitis B virus, hepatitis E virus (1) and others. These antibodies are valuable tools for investigating antigenic structure of viruses (2), development of diagnostic assays and prevalence studies of viral infections. Virus research is carried out in collaboration with Prof. Dr. R.Ulrich (Friedrich-Loeffler-Institute, Greifswald, Insel-Riems, Germany), Prof. Dr. D. Glebe (Giessen University, Germany), J.O.Koskinen (ArcDia International Oy Ltd, Turku, Finland) and other partners. We have also generated a collection of antibodies against bacterial cytolysins and exploited them both for structural studies and quantitation of cytolysins (3). In collaboration with our colleagues from the Department of Eukaryote gene engineering, we have developed a new technology for the use of virus-like particles as a carrier for target epitopes to increase their immunogenicity. This approach provides possibilities to generate antibodies against short and non-immunogenic protein sequences. For construction of recombinant antibodies, gene sequences encoding the variable parts of immunoglobulin heavy and light chains are cloned from hybridoma cells producing well-characterized monoclonal antibodies against the target of interest. Recombinant antibodies are developed in different formats - as single chain antibodies (scFv) and Fc-engineered antibodies where the scFv derived from hybridoma cells are joined to human IgG Fc fragment. Also, we have exploited recombinant virus-like particles as a carrier for antibody molecules, both scFv and Fc-engineered scFv. This innovative approach allows generation of recombinant multimeric antibodies displayed on virus-like particles as demonstrated for vaginolysin- specific antibodies and neutralizing antibodies against hepatitis B virus (4).
Main publications
- Simanavicius et al. Generation in yeast and antigenic characterization of hepatitis E virus capsid protein virus-like particles. Appl Microbiol Biotechnol., 2018, 102 (1), 185-198.
- Kailasan et al. Mapping antigenic epitopes on the human bocavirus capsid. J Virol., 2016, 90 (9), 4670-4680
- Zilnyte et al. The cytolytic activity of vaginolysin strictly depends on cholesterol and is potentiated by human CD59. Toxins (Basel). 2015, 7(1) :110-128
- Pleckaityte et al. Construction of polyomavirus-derived pseudotype virus-like particles displaying a functionally active neutralizing antibody against hepatitis B virus surface antigen. BMC Biotechnol., 2015, 15 (1): 85.






